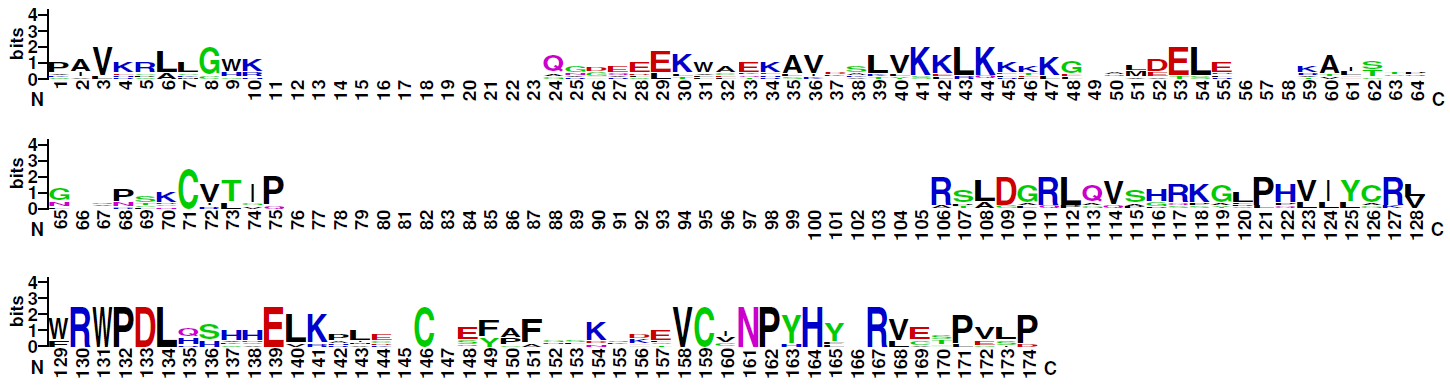| Description: |
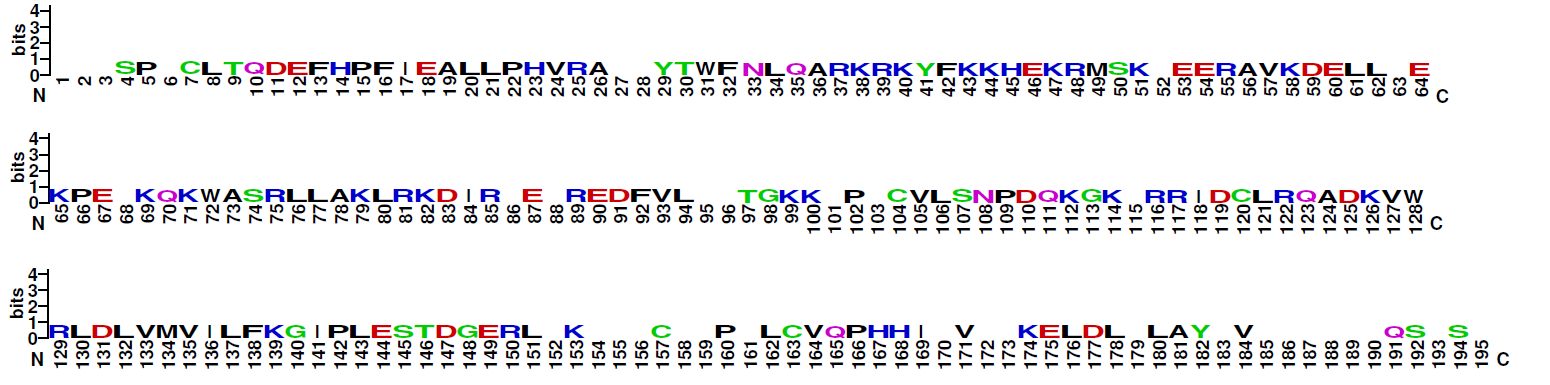
TRANSFAC® class description C0059: Significant similarities between DNA binding domains of Smad and NF1 factors have been revealed by a PSI-BLAST database search. Authors have proposed that Smad and NF1 factors belong to the same group of genes probably resulting from the common ancestry [25470]. For more details about Smad or NF1 factors see and correspondingly. TRANSFAC class description C0041: Proteins with 2 MAD-homology domains at N- and C-terminal ends, which have been named MH1 and MH2, are independently folded structures (PMID 10605817); the structure of the MH2 domain is a beta-sandwich with a 3-alpha-helix bundle at one end, and 3 large loops and an alpha-helix at the other end (PMID 10605817); the loop-helix region of one subunit packs against the 3-alpha-helix bundle of another subunit (PMID 10605817); MH1, a novel type of DBD, consists of 4 alpha-helixes, 6 short beta-strands, and 5 loops. Beta-strands form 2 small beta-sheets and 1 beta-hairpin (PMID 9741623); base-specific DNA recognition of the MH1 domain bound to a GTCT motif is via an 11-amino acid beta-hairpin, which lies in the major groove of the DNA (PMID 9741623, PMID 10605817); 3 classes of SMAD: Inhibitory SMADs (SMAD-6,SMAD-7), Receptor-regulated SMADs (SMAD-1, SMAD-2, SMAD-3, SMAD-5, SMAD-8), Common Mediator SMADs (SMAD-4, SMAD-4beta, SMAD-10). (PMID 10605817) |